Motivation
I live in Berlin, which is fast becoming the tech hub of Europe. Over the past two years we have seen a massive up-tick in the number of healthcare oriented startups, which are receiving funding on the basis of their use of Artificial Intelligence (AI). As somebody who knows a bit about the underlying technical and application problems of AI, what I see has made me very uneasy.
As a result of some personal conversations I was invited to give a Keynote address to the Digital Health Forum of the Berlin Institute of Health in March. This is a big deal because the BIH is a joint venture between Europe’s biggest teaching hospital, Charité Berlin, and one of Germany’s foremost centres for biological research, the Max-Delbrueck-Center, Berlin. The talk was extremely well received, so I have now given a public version at PyData Berlin 2018 which will be published on their YouTube channel in the coming weeks. (Update: the video is available here). In the meantime, I have written the discussion up as an article in two parts: part I is below and part II is available here.
Introduction
The world of biological modelling is broadly-speaking broken into two very distinct camps: statistical modellers and biophysicists. I am unusual in that I have worked in both camps. The purpose of statistically-based techniques is to gather correlative evidence, based on large sample sizes. Biophysical techniques, on the other hand, attempt to build knowledge of underlying processes into a model in order to i) better predict outcomes, or ii) find evidence for an alternative underlying physical explanation.
Today, I am largely interested in comparing data-driven machine learning techniques, which are a set of approaches which see growing use, with bespoke bottom-up pure modelling approaches.
When we think about AI we have to recognise that the term is poorly defined. The current use of the word, particularly where it pertains to applications, is in statistical or data-driven approaches. CEOs who are promising AI-driven approaches typically mean that they will use a black-box machine learning method and apply it to some data set.
I will look at both positives and negatives of such approaches, and then compare them with the bespoke approaches which I espoused for much of my career, before arriving at a synthesis1 which will hopefully be more powerful going forward. Regardless of which approach you choose to follow, it helps to know the limitations of your toolset.
The power of data-driven approaches
I don’t have to say too much to encourage people about the potential power of data-driven approaches today. Every time you hear a news story about AI driving some new application, you can be pretty sure that it involves one of these approaches.
In my opinion, computers are at their most powerful when they are providing decision support to expert human operators. In the case of medicine this means relieving the medic of many of the smaller choices (including via encumbering interfaces or paper procedures) which tire them as their shift progresses.
Many of the choices which a doctor makes during their day can be automated, these are the easy cases. By forcing a doctor to engage in many small-scale choices we reduce their ability to make bigger decisions as the day wears on. Numerous studies have described a phenomenon known as decision fatigue, and the results are particularly worrisome in clinical contexts.
Initial attempts in clinical support are in the area of electronic patient records. Without these, more advanced (AI-based) solutions cannot be built. By having all information at the fingertips of the doctor and combining this with a lightweight interface which hides all but the most pertinent data, we hope to enable doctors to perform at their peak decision making ability for longer proportions of their shifts.
Automated diagnosis and treatment systems have been on the horizon for a very long time. Working in machine learning, in the early 2000s, the accepted wisdom was that decision tree methods had already been proven to outperform medical practitioners at diagnosis
Any comparison of computer and human performance is highly dependent on how you choose your metric. Somebody needs to choose how to enter the symptoms into the computer, etc. But there is no doubt, at this point, that computers are much better at following a standardised diagnosis and treatment protocol, especially in the case of rare diseases, than human medics. Much like in the training of modern pilots, the training of medics is largely about getting them to follow standardised diagnostic protocols rather than building their individual approaches. The practice of medicine has improved as more and more doctors follow the successful treatment patterns laid down by the leaders in the field (evidence-based medicine) rather than plowing their own furrows.
However, compared to computers, human medics are much more effective at inferring what a patient is really presenting for, which may not be what they claim in their admission form. They also notice other symptoms which the patient may not have thought were relevant enough to mention. I am aware of a number of ongoing trials where automated systems are being connected, on the backend, to electronic patient records in order to catch errors and relieve burden from doctors.
Automation of image classification in the clinical context is something which I am particularly excited about. Currently, when you undergo a ‘scan’ a radiographer needs to look at the image in order to check it for a whole host of risk factors quite apart from the issue for which you were sent to them. On a daily basis each radiographer looks at 100s if not 1000s of scans. This is highly repetitive and quite tiring. Work is now well underway to automate much of this practice. The automation will not replace the radiographer but rather augment them, hopefully both simplifying the easy cases with salient feature highlighting and also catching the outliers which might have been overlooked.
There are numerous such approaches at the moment. I did a quick Google search in January and used the example of ChemXNet from Andrew Ng’s lab in my talk. More recently there was a Slashdot article about a similar system called BioMind which was developed in collaboration with Beijing’s Tiantan Hospital. BioMind apparently performs at expert-level in diagnosing brain tumours and haematoma expansion from images and does so in half the time a human requires.
These are the good news stories of the use of AI in Healthcare. So why should we think twice before throwing all of our R&D budgets at this field?
The limitations of data-driven approaches when applied to biology
I take the following as my starting point when I think about these methods:
Big data approaches are methods for finding statistical correlations in high dimensional (D) data taking advantage of large sample size (N).
There are many specific techniques for getting around issues of having lower-dimensional datasets or less data-points. But let’s be clear, specific advanced techniques are not the focus of the current trends in AI. We’re basically looking at Deep Neural Networks and their ilk here.
As someone who has worked with machine learning techniques, but who also has considerable experience working on biological applications, I see 5 major limitations of using such techniques in biology.
Limitation #1
Users often mistake N and D when looking at biological data.
In biology, D is often high but not necessarily informative. This breaks down in a number of ways. For example, the dimensionality of interactions in my body is almost uncountably large, but we only have access to a few of these dimensions. More importantly, the dimensions (parameters) to which we have access are not necessarily orthogonal to one another, so informational gains by adding parameters are typically quite small.
A simple example of this lack of informativeness in adding channels is in the case of the recent (2018) influenza outbreak in Berlin. If you presented at your doctor during this period with even a single symptom which correlated with influenza most doctors were immediately ready to diagnose. This contrasts with normal years, particularly here in Germany, where blood tests are required to confirm such a diagnosis. Now, adding data points about sweating, elevated temperature, blood pressure and heart rate are not really helping the diagnosis much since these are all correlated with a state of ill-health but not uniquely identifying with influenza.
Meanwhile, N is typically severely restricted in biology. In my original field of expertise, neuroscience, we typically dealt with sample-sizes of between 3 and 13. In larger-scale behavioural studies researchers typically fail to go much higher than 300 participants. This is perfectly adequate for a scientific study which is correctly analysed using appropriate statistical techniques. But it is nowhere near enough data points to fit a modern machine learning model.
When you learn about data driven techniques, the actual practice of the field is to try and turn everything into a question of, “How can I get more data?” This is an excellent engineering approach to using a massively effective tool. It works particularly well on website hits, where you just need to get more visitors in order to build a better service. It also works well on genome data: I do not want to claim that such techniques should never be used for biological applications! But there is a reason it has not been rolled out wider across biology. It’s just way too costly/labour intensive and the potential returns are not high enough; don’t forget that you would need to do massive-scale studies for each of the desired applications.
The final issue about N and D when it comes to biology is that many practitioners (PhD students, early-stage team members) often mistake N and D. In an attempt to increase N practitioners often inappropriately slice the data. The best example I can give is in handling time-series data. There are cases where each data-point in a time-series can be handled as a separate data-point, but they are few and need to be motivated. But in early-stage analyses researchers often postpone the motivation part and just treat all of the data-points separately. This certainly boosts N, but it is deeply skewing the training data set by effectively artificially resampling from the same small number of individuals 1000s of times.
Limitation #2
In repeated testing in high D you will always find correlations
If you don’t understand this point then I refer you to the excellent website Spurious Correlations. There is even a book!
I will write a technical article on this topic at another time. But the basic idea is that even independent data sources can appear correlated for periods. They are independent, so the observation of one cannot influence the other. Now, if you gather enough data from two independent sources then you will find arbitrarily long sub-sequences for which they are strongly correlated.
If instead you go to high dimensionality (D) you have very many data sources. In this case you no longer need to gather incredibly large data sets, you just need to look across the pairings of dimensions and you will find two that have a given level of correlation for the entire data set.
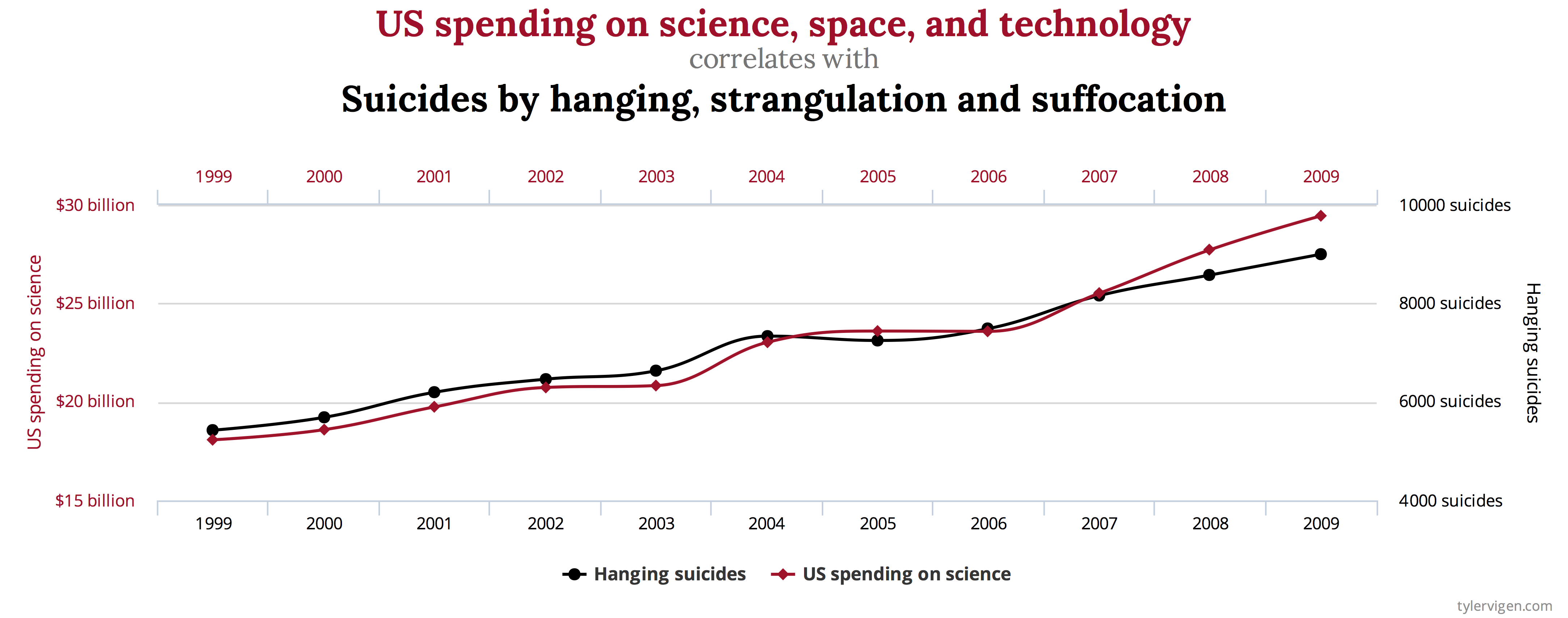
The whole theory of statistics and sampling is based around this concept. What is at issue is that these correlations are spurious (or may be), represent epiphenomena and are not predictive of the actual system behaviour.
Limitation #3
Data driven techniques have a historical ideological preference for model-free approaches
These techniques were born in the 1980s and 1990s and were in some senses a response to trends in Statistics and other fields. The idea was to set-aside preconceptions of how data should look and to develop generically applicable techniques.
History has shown this to be enormously successful. Particularly, since Google discovered that by throwing 100x more data at the models you can develop solutions which are actually better than hand-constructed methods. Artisanal approaches are costly and often miss something, but given enough (cheap) data the machine learning approach can, in theory, capture everything.
However, given that Limitation #1 suggests that the gathering of 100x more data may be difficult and costly in biology we need to appreciate that a model-free approach may be restricting. Biology is largely a deterministic process. There are many elements which we do not understand, and some theories suggest that at a molecular interaction level randomness may be a feature, but our best guess is that mechanism and function in biology are strongly deterministic. So, why are we ignoring these mechanisms in our applications?
Furthermore, biological data can be extremely noisy. Technical limitations lead to poor recordings and it is difficult to isolate processes. When you are dealing with noisy data, especially with small sample size (N), one very effective way of reducing the signal-to-noise ratio is the use of a model.
Limitation #4
Don’t confuse generalization with generalisation
In the machine learning world, generalization is the ability to treat statistically identical data. It does not imply an ability to deal with data points outside of the original training set.
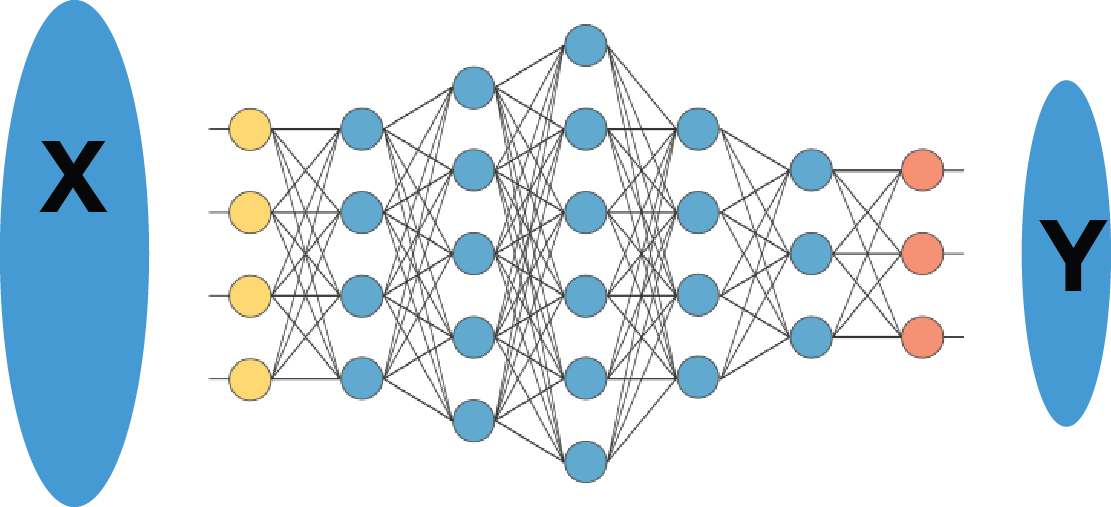
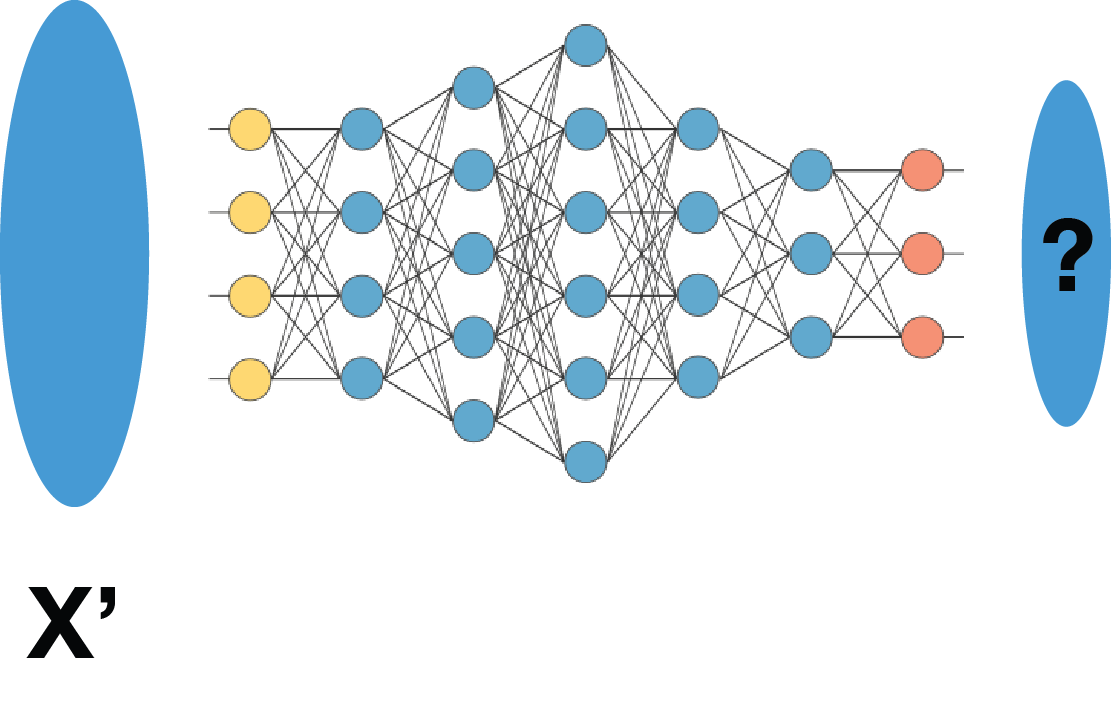
Just to confuse things even further, techniques such as Deep Neural Networks have not yet been shown to map data points onto a smoothly differentiable manifold. This is a technical point, and I am aware of attempts to overcome it. But the basic meaning is that two cases which we might think should be almost identical may not be represented in almost identical ways in the network, nor be mapped to almost identical outputs.
Limitation #5
Data driven techniques cannot identify mechanism
Our understanding of the world is mechanistic. I am currently reading Judea Pearl’s opus The Book of Why. In it, I recognise a description of the type of mathematics which I have used intuitively for much of my career. Pearl’s theory is that we rationalise the world in terms of Why? This is extremely powerful and potentially at the root of our intelligence.
Medical doctors are willing to ignore questions of Why on the basis that their first priority is to treat their patients. But science, including medical science, does not progress very far in the absence of theories about function.
Remember, data-driven techniques only really work with data points which are statistically identical to points which they have seen before. This means that they cannot really generalise new information or suggest novel treatments. You can trick a few suggestions out of them, which might work on some early cases, but this approach quickly stalls.
In the absence of a theory of mechanism these techniques are borderline useless for developing new theories or testing novel ideas. Biology, in particular, is completely bound up in mechanism so this is a major limiting issue.
Continued in Part II
Part II of this article, in which I look at biophysical modelling approaches and finish up with hybrid solutions.

Great article…… great clarity! Thanks for the reference to Pearl‘s work also! Mechanism and causality……..